Decoding the regulatory role of non-coding genome sequence.
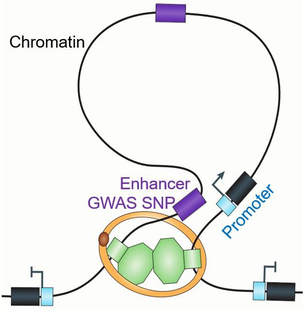
More than 98% of the human genome are non-coding sequence and vast majority (~93%) of human disease and traits associated risk variants lie in these non-coding regions. These non-coding regions, that were previously considered as non-essential and called as “junk DNA” or “dark matter” in the genome, are now recognized as important for controlling spatiotemporal gene expression. A growing body of evidence indicates in many cases that the genetic and epigenetic variants underlying the non-coding regulatory sequences, such as enhancer, insulator, and repressive elements, contribute to human diseases. Yet, we know little about the consequences and mechanisms of how cis-regulatory element and disease-associated non-coding variations regulate development, tissue regeneration, degenerative disorders, and cancer. Our lab will develop and apply innovative high-throughput functional genomics and genome editing tools that allow us to investigate how the “junk DNA” interplay with transcription factors and epigenetic mechanism to control gene regulation.
More than 98% of the human genome are non-coding sequence and vast majority (~93%) of human disease and traits associated risk variants lie in these non-coding regions. These non-coding regions, that were previously considered as non-essential and called as “junk DNA” or “dark matter” in the genome, are now recognized as important for controlling spatiotemporal gene expression. A growing body of evidence indicates in many cases that the genetic and epigenetic variants underlying the non-coding regulatory sequences, such as enhancer, insulator, and repressive elements, contribute to human diseases. Yet, we know little about the consequences and mechanisms of how cis-regulatory element and disease-associated non-coding variations regulate development, tissue regeneration, degenerative disorders, and cancer. Our lab will develop and apply innovative high-throughput functional genomics and genome editing tools that allow us to investigate how the “junk DNA” interplay with transcription factors and epigenetic mechanism to control gene regulation.