1. Developing 'omics technologies to study chromatin structure and function in the context of gene regulation.
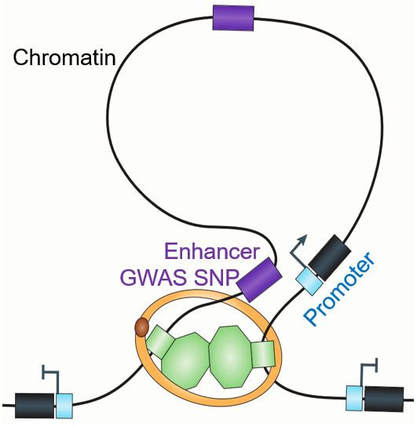
Less than 2% of the genome sequences are protein coding genes, while more than 98% of the human genome are non-coding sequence. Surprisingly, vast majority (~93%) of genetic variants that are associated with human traits and diseases lie in the non-coding genomic regions. When the human genome was first sequenced, these non-coding sequences were considered as non-essential and called as “junk DNA” or “dark matter”. However, accumulating evidence showed that these non-coding sequences can be epigenetically modified and bound by transcription factors to exert a critical role in regulating temporal-spatial gene expression program. Genetic and epigenetic alterations on these non-coding regulatory elements, such as promoter, enhancer, insulator, and silencer, often leads to human diseases. Yet, we still know little about the structure, function, regulation and biology of non-coding regulatory genome. We have a track record of developing new high-throughput genomics tools to further our understanding of chromatin biology. On this research direction, we are currently focused on the following topics: CRISPR-BioID; single-cell multi-omics profiling and perturbation; 3D genome analysis; and in vivo genome/epigenome engineering. These projects are funded by NHGRI Genome Innovator Awards, and the 4D Nucleome Consortium program by NIH Director's Office.
2. Gene regulation of skeletal muscle regeneration.
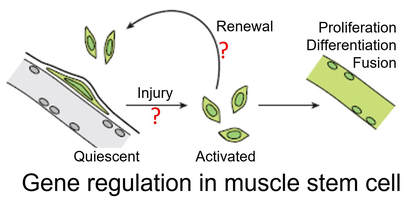
Muscle stem cell (MuSC), also known as satellite cell, is the major cell type that directly contributes to skeletal muscle development, growth and repair. In a broad spectrum of pathologic conditions, such as muscular dystrophy, aging, ischemia, and cachexia (devastating weight loss as well as muscle wasting associate with chronic conditions), accumulation of stem cell-intrinsic damages as well as the deleterious “niche” lead to a marked decline of muscle stem cell repair ability. MuSC transplantation offers great promise to treat muscular disorders, but its application has been hindered by a lack of understanding of the gene regulation mechanism that preserve stem cell long-term regenerative potency. Our group will strive to expand the understanding of the transcriptional and epigenetic control mechanism determining muscle stem cell quiescence, "alert", activation, self-renewal, and cell fate determination in health, disease, and aging. In the long run, we hope to translate the knowledge into innovative stem-cell based therapies to treat muscular disorders. These projects are support by NIH 4D Nucleome Consortium, American Federation for Aging Research (AFAR) and Glenn Foundation for Medical Research.
3. Gene regulation of rhabdomyosarcoma tumorigenesis.
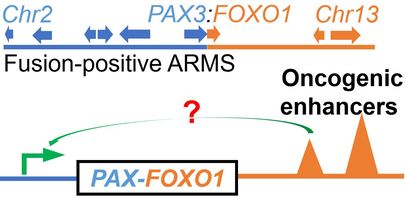
Disruption of gene expression program causes diseases, including cancer. Rhabdomyosarcoma (RMS) is a life-threatening pediatric cancer often regarded as "myogenesis go awry" with imbalanced myogenic proliferation and differentiation program. Therefore, we hypothesize that the normal gene regulation mechanism that are needed for muscle repair are deregulated, by genetic or epigenetic alterations, in the RMS cells. We combine our expertise on myogenesis, high-throughput genomics analysis, CRISPR-BioID, and high-throughput CRISPR perturbation assays, in genetically engineered mouse model and human patient-derived RMS sphere culture, to generate data, analyze data, and formulate hypothesis from the data. We are closely collaborate with scientists and physicians at Duke and across the country to further our understanding of this deadly pediatric cancer. This research direction has been supported by V Scholar for Cancer Research, the Elsa U. Pardee Foundation, and Cancer Moonshot FusOnC2 Consortium.
Selected Publications:
(* co-first author; # co-corresponding author)
Complete List of Published Work also available at: https://www.ncbi.nlm.nih.gov/myncbi/yarui.diao.1/bibliography/public/
and Google Scholar
Yarui Diao's key publications as a trainee:
1. Diao Y, Wang X, Wu Z. SOCS1, SOCS3, and PIAS1 promote myogenic differentiation by inhibiting the leukemia inhibitory factor-induced JAK1/STAT1/STAT3 pathway. Mol Cell Biol. 2009 Sep;29(18):5084–5093. PMCID: PMC2738280
2. Diao Y, Liu W, Wong CCL, Wang X, Lee K, Cheung PY, Pan L, Xu T, Han J, Yates JR 3rd, Zhang M, Wu Z. Oxidation-induced intramolecular disulfide bond inactivates mitogen-activated protein kinase kinase 6 by inhibiting ATP binding. Proc Natl Acad Sci U S A. 2010 Dec 7;107(49):20974–20979. PMCID: PMC3000308
3. Diao Y, Guo X, Li Y, Sun K, Lu L, Jiang L, Fu X, Zhu H, Sun H, Wang H, Wu Z. Pax3/7BP is a Pax7- and Pax3-binding protein that regulates the proliferation of muscle precursor cells by an epigenetic mechanism. Cell Stem Cell. 2012 Aug 3;11(2):231–241. PMID: 22862948
4. Diao Y, Guo X, Jiang L, Wang G, Zhang C, Wan J, Jin Y, Wu Z. miR-203, a tumor suppressor frequently down-regulated by promoter hypermethylation in rhabdomyosarcoma. J Biol Chem. 2014 Jan 3;289(1):529–539. PMCID: PMC3879574
5. Diao Y, Li B, Meng Z, Jung I, Lee AY, Dixon J, Maliskova L, Guan KL, Shen Y, Ren B. A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Res. 2016 Mar;26(3):397–405. PMCID: PMC4772021
6. An Y*, Wang G*, Diao Y*, Long Y, Fu X, Weng M, Zhou L, Sun K, Cheung TH, Ip NY, Sun H, Wang H, Wu Z. A Molecular Switch Regulating Cell Fate Choice between Muscle Progenitor Cells and Brown Adipocytes. Dev Cell. 2017 May 22;41(4):382–391.e5. PMID: 28535373
7. Diao Y, Fang R, Li B, Meng Z, Yu J, Qiu Y, Lin KC, Huang H, Liu T, Marina RJ, Jung I, Shen Y, Guan KL, Ren B. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat Methods. 2017 Jun;14(6):629–635. PMCID: PMC5490986
8. Jung I*, Schmitt A*, Diao Y*, Lee AJ, Liu T, Yang D, Tan C, Eom J, Chan M, Chee S, Chiang Z, Kim C, Masliah E, Barr CL, Li B, Kuan S, Kim D, Ren B. A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat Genet. 2019 Oct;51(10):1442–1449. PMCID: PMC6778519
Yarui Diao's key publications as a PI:
9. Chen X, Wan J, Yu B, Diao Y#, Zhang W#. PIP5K1α promotes myogenic differentiation via AKT activation and calcium release. Stem Cell Res Ther. 2018 9;9(1):33. PMCID: PMC5806439
10. Li B, Chen PB, Diao Y. CRISPR-SE: a brute force search engine for CRISPR design. NAR Genom Bioinform. 2021 Mar;3(1):lqab013. PMCID: PMC7902234
11. Wei X, Xiang Y, Peters DT, Marius C, Sun T, Shan R, Ou J, Lin X, Yue F, Li W, Southerland KW, Diao Y. HiCAR is a robust and sensitive method to analyze open-chromatin-associated genome organization. Mol Cell. 2022 Mar 17;82(6):1225–1238.e6. PMCID: PMC8934281
12. Sun T, Xu Y, Xiang Y, Ou J, Soderblom EJ, Diao Y. Crosstalk between RNA m6A and DNA methylation regulates transposable element chromatin activation and cell fate in human pluripotent stem cells. Nat Genet. 2023 Aug;55(8):1324–1335. PMCID: PMC10766344
13. Okafor AE, Lin X, Situ C, Wei X, Xiang Y, Wei X, Wu Z, Diao Y. Single-cell chromatin accessibility profiling reveals a self-renewing muscle satellite cell state. J Cell Biol. 2023 Aug 7;222(8). PMCID: PMC10309185
14. Wei X, Tran D, Diao Y. HiCAR: Analysis of Open Chromatin Associated Long-range Chromatin Interaction Using Low-Input Materials. Curr Protoc. 2023 Oct;3(10):e899. PMCID: PMC10575683
15. Southerland KW, Xu Y, Peters DT, Lin X, Wei X, Xiang Y, Fei K, Olivere LA, Morowitz JM, Otto J, Dai Q, Kontos CD, Diao Y. Skeletal muscle regeneration failure in ischemic-damaged limbs is associated with pro-inflammatory macrophages and premature differentiation of satellite cells. Genome Med. 2023 Nov 10;15(1):95. PMCID: PMC10636829